systemPipeR and CWL
Last update: 06 August, 2021
cwl_and_spr.RmdAuthors: Daniela Cassol (danielac@ucr.edu), Le Zhang (le.zhang001@email.ucr.edu), Thomas Girke (thomas.girke@ucr.edu).
Institution: Institute for Integrative Genome Biology, University of California, Riverside, California, USA.
How to connect CWL description files within systemPipeR
This section will demonstrate how to connect CWL parameters files to create workflows. In addition, we will show how the workflow can be easily scalable with systemPipeR.
SYSargsList container stores all the information and instructions needed for processing a set of input files with a single or many command-line steps within a workflow (i.e. several components of the software or several independent software tools). The SYSargsList object is created and fully populated with the SYSargsList construct function. Full documentation of SYSargsList management instances can be found here.
The following imports a .cwl file (here example.cwl) for running the echo Hello World! example.
HW <- SPRproject(projPath = tempdir())
#> Creating directory: /tmp/RtmpzS70mx/data
#> Creating directory: /tmp/RtmpzS70mx/param
#> Creating directory: /tmp/RtmpzS70mx/results
#> Creating directory '/tmp/RtmpzS70mx/.SPRproject'
#> Creating file '/tmp/RtmpzS70mx/.SPRproject/SYSargsList.yml'
#> Your current working directory is different from the directory chosen for the Project Workflow.
#> For accurate location of the files and running the Workflow, please set the working directory to
#> 'setwd(/tmp/RtmpzS70mx)'
HW <- SYSargsList(wf_file = "example/workflow_example.cwl",
input_file = "example/example_single.yml",
dir_path = system.file("extdata/cwl", package = "systemPipeR"))
HW
#> Instance of 'SYSargsList':
#> WF Steps:
#> 1. Step_x --> Status: Pending
#> Total Files: 1 | Existing: 0 | Missing: 1
#> 1.1. echo
#> cmdlist: 1 | Pending: 1
#>
cmdlist(HW)
#> $Step_x
#> $Step_x$defaultid
#> $Step_x$defaultid$echo
#> [1] "echo Hello World! > results/M1.txt"However, we are limited to run just one command-line or one sample in this example. To scale the command-line over many samples, a simple solution offered by systemPipeR is to provide a variable for each of the parameters that we want to run with multiple samples.
Let’s explore the example:
dir_path <- system.file("extdata/cwl", package = "systemPipeR")
yml <- yaml::read_yaml(file.path(dir_path, "example/example.yml"))
yml
#> $message
#> [1] "_STRING_"
#>
#> $SampleName
#> [1] "_SAMPLE_"
#>
#> $results_path
#> $results_path$class
#> [1] "Directory"
#>
#> $results_path$path
#> [1] "./results"For the message and SampleName parameter, we are passing a variable connecting with a third file called targets.
Now, let’s explore the targets file structure:
targetspath <- system.file("extdata/cwl/example/targets_example.txt", package = "systemPipeR")
read.delim(targetspath, comment.char = "#")
#> Message SampleName
#> 1 Hello World! M1
#> 2 Hello USA! M2
#> 3 Hello Bioconductor! M3The targets file defines all input files or values and sample ids of an analysis workflow. For this example, we have defined a string message for the echo command-line tool, in the first column that will be evaluated, and the second column is the SampleName id for each one of the messages. Any number of additional columns can be added as needed.
Users should note here, the usage of targets files is optional when using systemPipeR's new CWL interface. Since for organizing experimental variables targets files are extremely useful and user-friendly. Thus, we encourage users to keep using them.
How to connect the parameter files and targets file information?
The constructor function creates an SYSargsList S4 class object connecting three input files:
- CWL command-line specification file (
wf_fileargument); - Input variables (
input_fileargument); - Targets file (
targetsargument).
As demonstrated above, the latter is optional for workflow steps lacking input files. The connection between input variables (here defined by input_file argument) and the targets file are defined under the inputvars argument. A named vector is required, where each element name needs to match with column names in the targets file, and the value must match the names of the .yml variables. This is used to replace the CWL variable and construct all the command-line for that particular step.
The variable pattern _XXXX_ is used to distinguish CWL variables that target columns will replace. This pattern is recommended for consistency and easy identification but not enforced.
The following imports a .cwl file (same example demonstrated above) for running the echo Hello World example. However, now we are connecting the variable defined on the .yml file with the targets file inputs.
HW_mul <- SYSargsList(step_name = "echo",
targets=targetspath,
wf_file="example/workflow_example.cwl", input_file="example/example.yml",
dir_path = dir_path,
inputvars = c(Message = "_STRING_", SampleName = "_SAMPLE_"))
HW_mul
#> Instance of 'SYSargsList':
#> WF Steps:
#> 1. echo --> Status: Pending
#> Total Files: 3 | Existing: 0 | Missing: 3
#> 1.1. echo
#> cmdlist: 3 | Pending: 3
#>
cmdlist(HW_mul)
#> $echo
#> $echo$M1
#> $echo$M1$echo
#> [1] "echo Hello World! > results/M1.txt"
#>
#>
#> $echo$M2
#> $echo$M2$echo
#> [1] "echo Hello USA! > results/M2.txt"
#>
#>
#> $echo$M3
#> $echo$M3$echo
#> [1] "echo Hello Bioconductor! > results/M3.txt"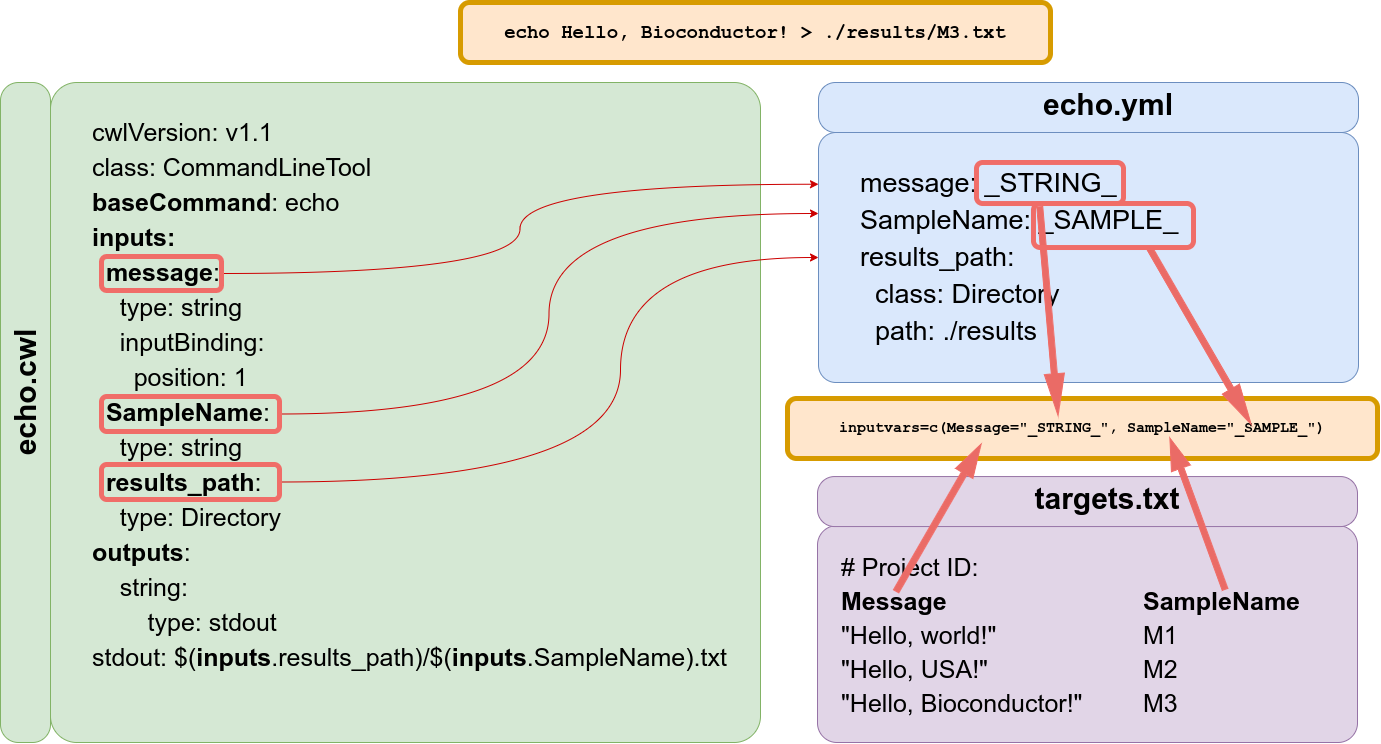
Connectivity between CWL param files and targets files.