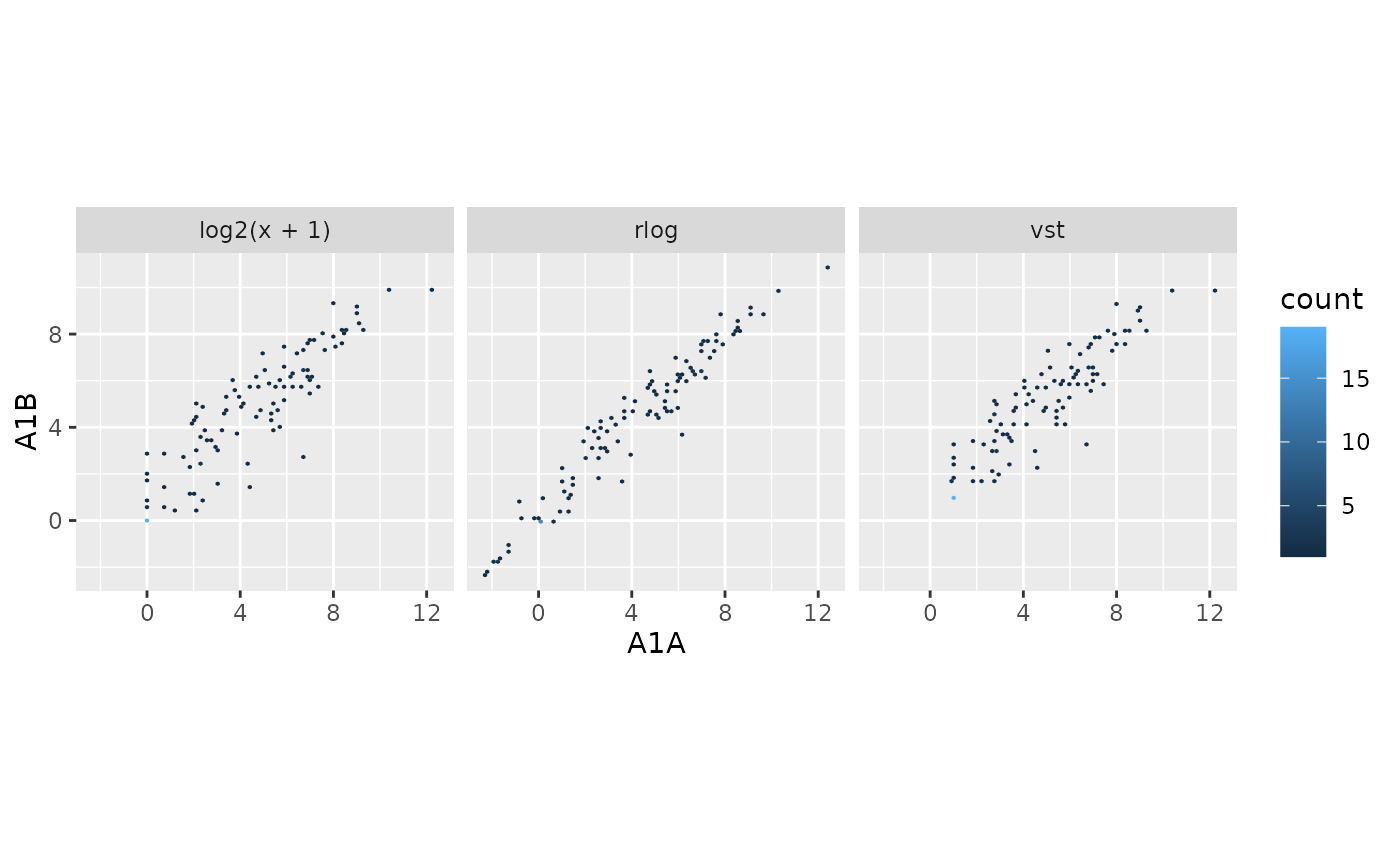
exploreDDSplot
exploreDDSplot.RdScatterplot of transformed counts from two samples or grid of all samples
exploreDDSplot( countMatrix, targets, cmp = cmp[[1]], preFilter = NULL, samples, blind = TRUE, scattermatrix = FALSE, plotly = FALSE, savePlot = FALSE, filePlot = NULL )
Arguments
| countMatrix |
|
|---|---|
| targets | targets |
| cmp |
|
| preFilter | allows removing rows in which there are very few reads.
Accepts a numeric value with the minimum of total reads to keep.
Default is |
| samples | a |
| blind | logical, whether to blind the transformation to the experimental
design (see varianceStabilizingTransformation), from |
| scattermatrix | if |
| plotly | logical: when |
| savePlot | logical: when |
| filePlot | file name where the plot will be saved. For more information,
please consult the |
Value
returns an object of ggplot2 plot.
Examples
## Targets file targetspath <- system.file("extdata", "targets.txt", package = "systemPipeR") targets <- read.delim(targetspath, comment = "#") cmp <- systemPipeR::readComp( file = targetspath, format = "matrix", delim = "-" ) ## Count table file countMatrixPath <- system.file("extdata", "countDFeByg.xls", package = "systemPipeR" ) countMatrix <- read.delim(countMatrixPath, row.names = 1) ## Plot exploreDDSplot(countMatrix, targets, cmp = cmp[[1]], preFilter = NULL, samples = c(3, 4) )