class: center, middle, inverse, title-slide # <p><img src="https://raw.githubusercontent.com/systemPipeR/systemPipeShiny/master/inst/app/www/img/sps_small.png" style="width:1in" /> <br/><em>systemPipeShiny</em></p> ### <br/>Daniela Cassol & Le Zhang ### <h3> University of California Riverside <h3/> ### <h3> 2020/08/28 (updated: 2020-08-31) <h3/> --- <!-- background-image: url(https://raw.githubusercontent.com/systemPipeR/systemPipeShiny/master/inst/app/www/img/sps_small.png) --> layout: true background-image: url(https://raw.githubusercontent.com/systemPipeR/systemPipeShiny/master/inst/app/www/img/sps_small.png) background-position: 100% 0% background-size: 10% --- class: center, middle ## Outline #### Introduction #### *systemPipeShiny* Demo #### Visualization Features #### Customize and Extend --- class: inverse, center, middle # Introduction --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities .pull-left[  ] .pull-right[ 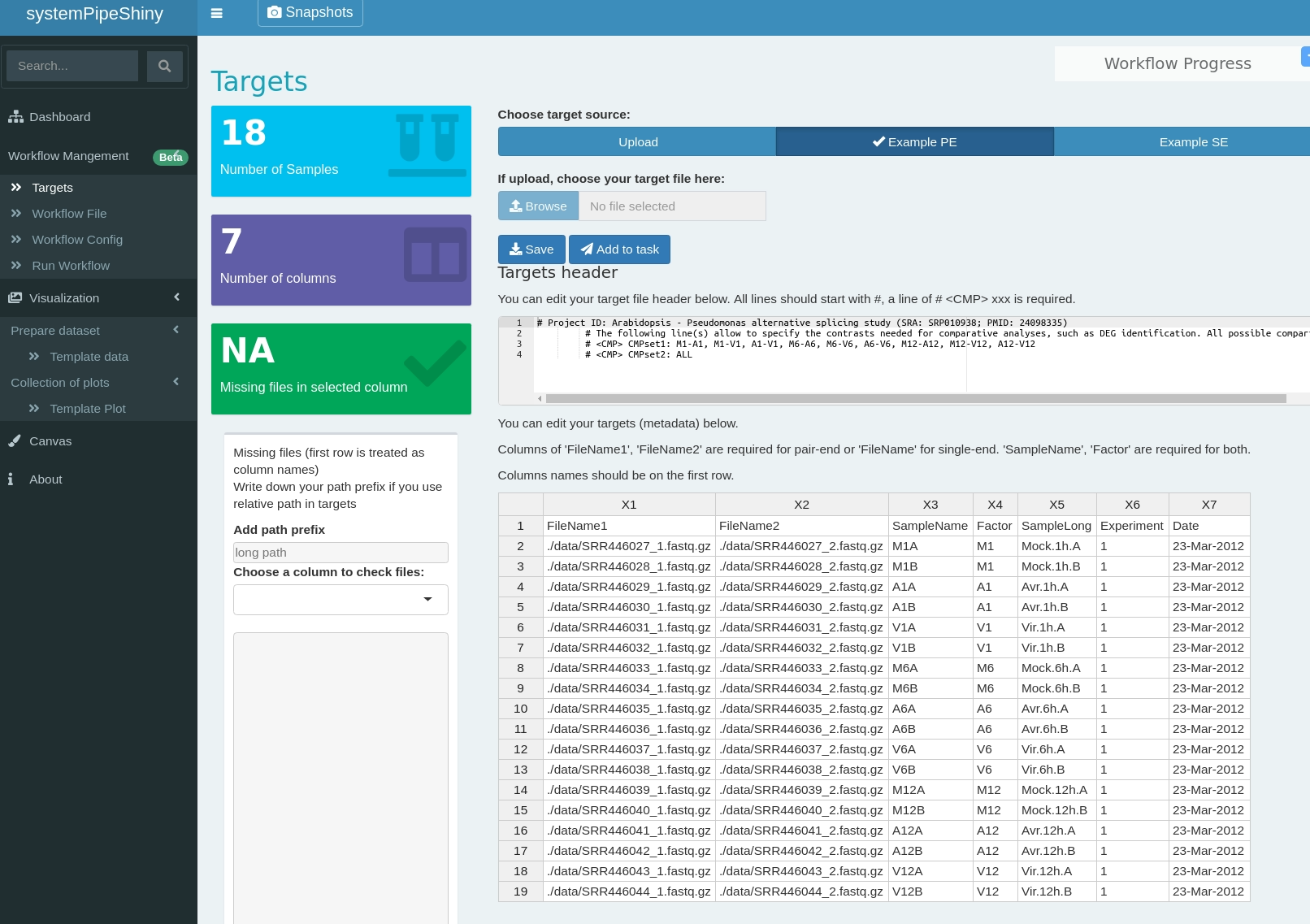 ] --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities .pull-left[  ] .pull-right[ 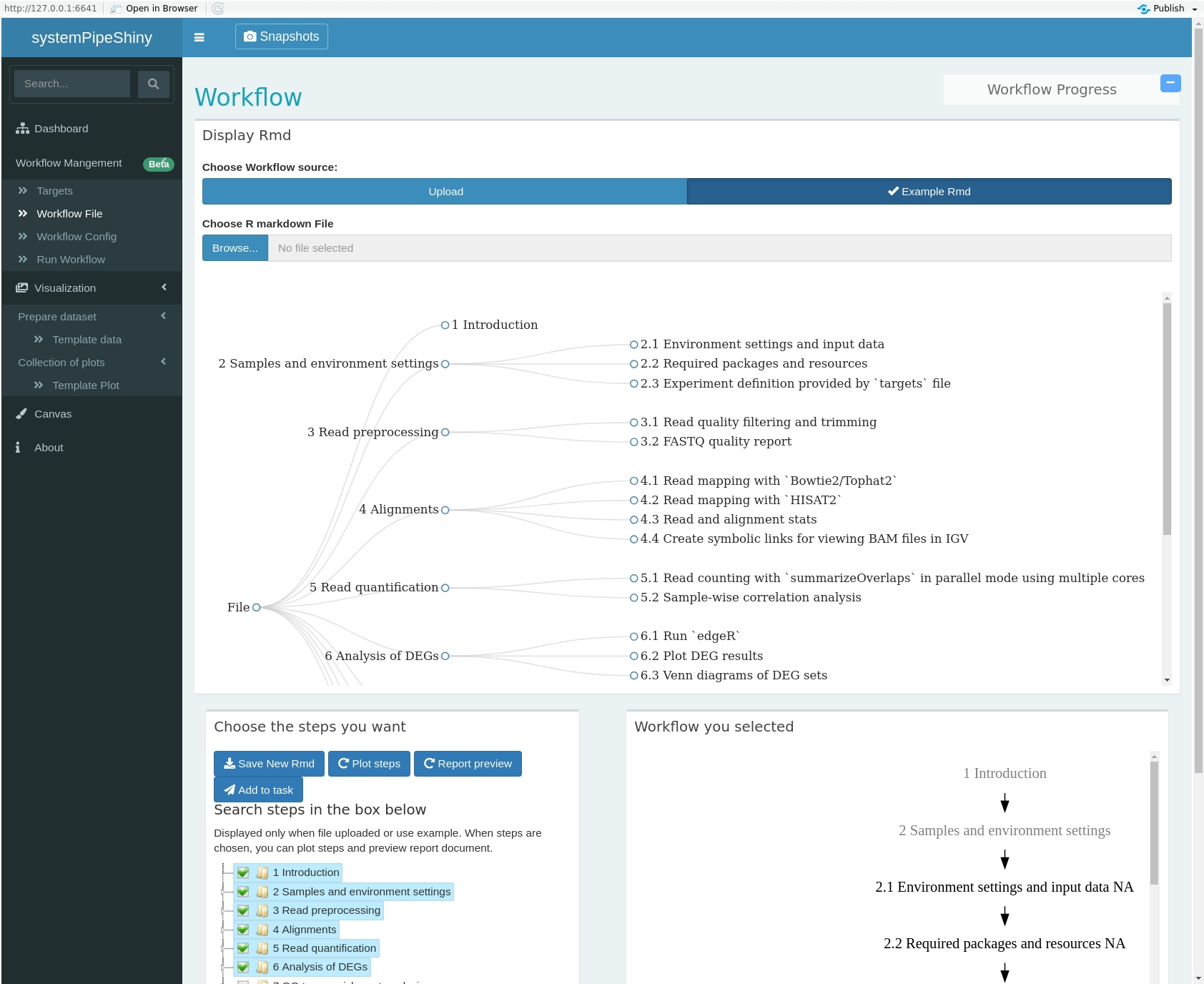 ] --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities .pull-left[ .center[  ]] .pull-right[ <a href="#add link"><img src="qual.png" width="190" height="440" title=targets alt=Targets></a> ] --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities .pull-left[  ] .pull-right[ <a href="#heatMap"><img src="heat_samples.svg"width="150" height="150" title=#heatMap alt=heat_samples.svg></a><a href="#cor"><img src="cor.svg"width="150" height="150" title=#cor alt=cor.svg></a><a href="#pca"><img src="tsne.svg"width="150" height="150" title=#pca alt=tsne.svg></a><a href="#volcano"><img src="volcano.svg"width="150" height="150" title=#volcano alt=volcano.svg></a><a href="#volcano"><img src="heat.png"width="150" height="150" title=#volcano alt=heat.png></a><a href="#logs"><img src="maplot.svg"width="150" height="150" title=#logs alt=maplot.svg></a> ] --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities .pull-left[  ] .pull-right[ 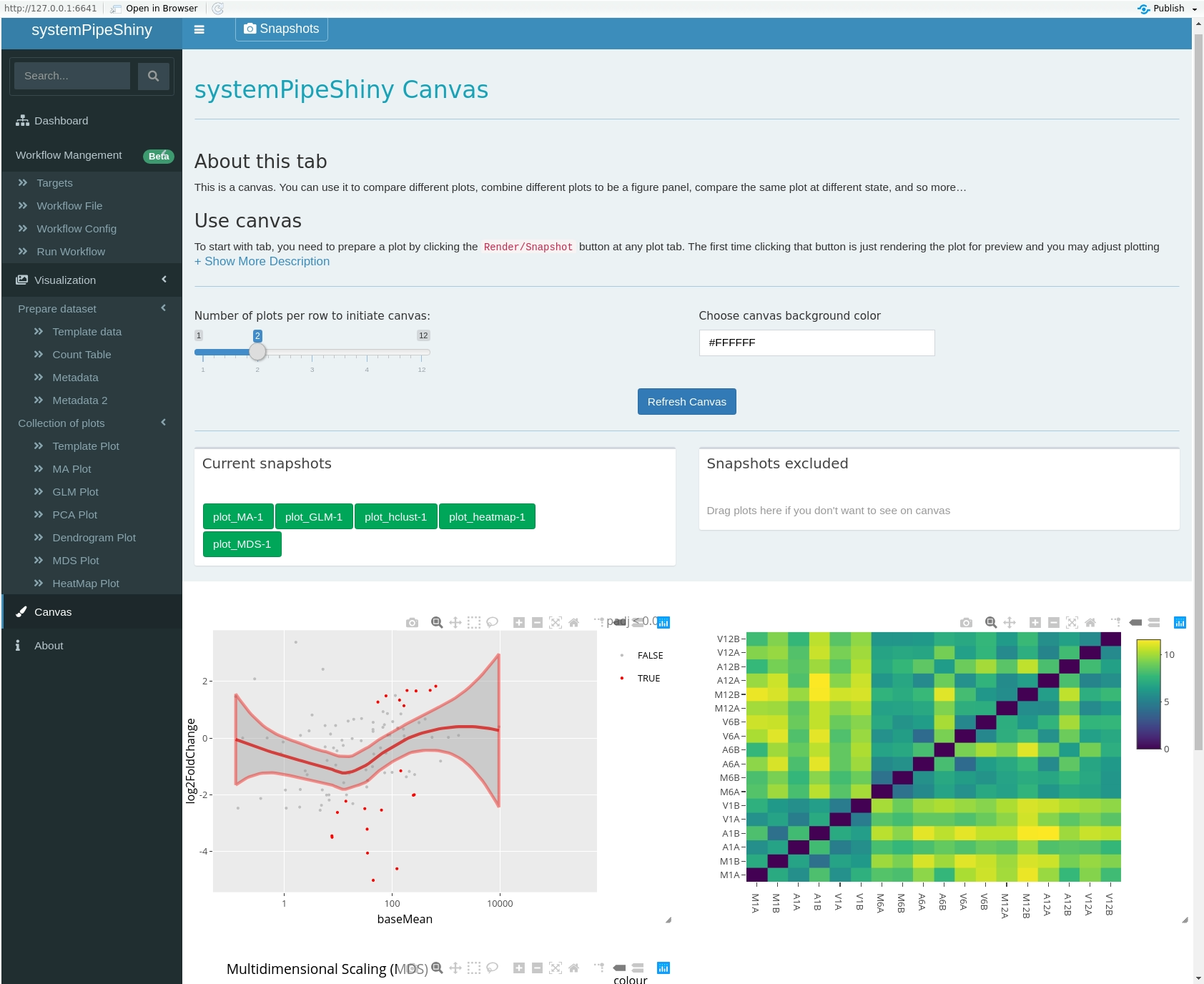 ] --- ## Motivation <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Build an interactive framework for workflow management and visualization by extending all [systemPipeR](https://systempipe.org/) functionalities <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Provide for non-R users, such as experimentalists, to run many systemPipeR’s workflow designs, control, and visualization functionalities interactively without requiring knowledge of R <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Provide a tool that can be used on both local computers as well as centralized server-based deployments that can be accessed remotely as a public web service for using SPR’s functionalities with the community and/or private data --- ## <i class="fas fa-toolbox"></i> Features .middle-slide[  ] ??? <i class="fas fa-wrench" style="color:#00758a;"></i> Interactively define experimental designs and provide associated metadata using an easy to use tabular editor and/or file uploader <i class="fas fa-wrench" style="color:#00758a;"></i> visualize workflow topologies combined with auto-generation of R Markdown previews for interactively designed workflows <i class="fas fa-wrench" style="color:#00758a;"></i> Allows prepare the data for the visualizations tabs --- ## <i class="fas fa-toolbox"></i> Structural Features -- .left-column[ ### User friendly ] .right-column[  ] --- ## <i class="fas fa-toolbox"></i> Structural Features .left-column[ ### User friendly ### Progress Tracking ] .right-column[  ] --- ## <i class="fas fa-toolbox"></i> Structural Features .left-column[ ### User friendly ### Progress Tracking ### Canvas ] .right-column[ Under this workbench users can take snapshots of different plots, and combine or resize them. This feature is useful for generating complex scientific summary graphics. <img src="canvas.png" alt="drawing" style="width:450px;"/> ] --- ## <i class="fas fa-toolbox"></i> Structural Features .left-column[ ### Progress Tracking ] .right-column[ Messages, warnings and errors from R functions are automatically captured and logged on both the server and client ends. <img src="error.png" alt="drawing" style="width:450px;"/> ] --- ## <i class="fas fa-toolbox"></i> Structural Features .left-column[ ### Progress Tracking ### App Config ] .right-column[ a robust exception handling system has been implemented (similar to Shiny options), that provides error solutions to to users, e.g. invalid parameter settings. ] --- ## <i class="fas fa-toolbox"></i> Structural Features .left-column[ ### Progress Tracking ### App Config ### Modular Design ] .right-column[ SPS is built on Shiny modules, which provides local scope isolation between each tab. Objects on one tab do not conflict with other tabs. To enable cross-tab communication, SPS also supports global scope interactions ] --- ## <i class="fas fa-bezier-curve"></i> Design  --- ## <i class="fas fa-user-edit"></i> Customize and Extend <i class="fas fa-wrench" style="color:#00758a;"></i> Templates are available to generate new visualization tabs  --- class: inverse, center, middle # <i class="fas fa-code"></i> Live Demo --- ## <i class="fas fa-box-open"></i> Install Package Install the **systemPipeShiny** package from [GitHub](https://github.com/systemPipeR/systemPipeShiny): ```r if (!requireNamespace("BiocManager", quietly=TRUE)) install.packages("BiocManager") BiocManager::install("systemPipeR/systemPipeShiny", dependencies=TRUE, build_vignettes=TRUE) ``` ### <i class="fas fa-book"></i> Load Package and Documentation <i class="fas fa-question" style="color:#00758a;"></i> Load packages and accessing help ```r library("systemPipeShiny") ``` <i class="fas fa-question" style="color:#00758a;"></i> Access help ```r library(help="systemPipeShiny") vignette("systemPipeShiny") ``` --- ## <i class="fas fa-code"></i> Quick Start Create the project: ```r systemPipeShiny::spsInit() ## [SPS-INFO] 2020-08-30 17:03:28 Start to create a new SPS project ## [SPS-INFO] 2020-08-30 17:03:28 Create project under /home/dcassol/danielac@ucr.edu/projects/Presentations/SPS/SPS_20200830 ## [SPS-INFO] 2020-08-30 17:03:28 Now copy files ## [SPS-INFO] 2020-08-30 17:03:28 Create SPS database ## [SPS-INFO] 2020-08-30 17:03:28 Created SPS database method container ## [SPS-INFO] 2020-08-30 17:03:28 Creating SPS db... ## [SPS-DANGER] 2020-08-30 17:03:28 Db created at '/home/dcassol/danielac@ucr.edu/projects/Presentations/SPS/SPS_20200830/config/sps.db'. DO NOT share this file with others ## [SPS-INFO] 2020-08-30 17:03:28 Key md5 dc17b12b7cadbb70e2d32a77bb32a17f ## [SPS-INFO] 2020-08-30 17:03:28 SPS project setup done! ``` --- ## <i class="far fa-folder-open"></i> SPS Folder Structure Directory structure: ``` SPS_YYYYMMDD ├── server.R ├── ui.R ├── global.R ## It will need manual input for new tabs ├── deploy.R ## Deploy helper file ├── config/ ## Folder with app configuration files │ ├── sps.db │ ├── sps_options.yaml │ └── tabs.csv ├── R/ ## All SPS additional tab files and helper R function files │ └── tabs_xx.R ├── data/ ## Storage all the input data │ └── inputData ├── results/ ## Storage all the results and plot data │ └── plot_xx.png └── www/ ## Folder with all the app resources ``` --- ## <i <i class="far fa-chart-bar"></i> Launching the interface The `runApp` function from `shiny` package launches the app in our browser. ```r shiny::runApp() ``` Check out our instance of **systemPipeShiny**: [Link](https://tgirke.shinyapps.io/systemPipeShiny/) Note: Add iframe here --- class: inverse, center, middle # <i class="fas fa-code"></i> Visualization Features --- ### <i class="far fa-chart-bar"></i> Data transformations and visualization .panelset[ .panel[.panel-name[Plot] 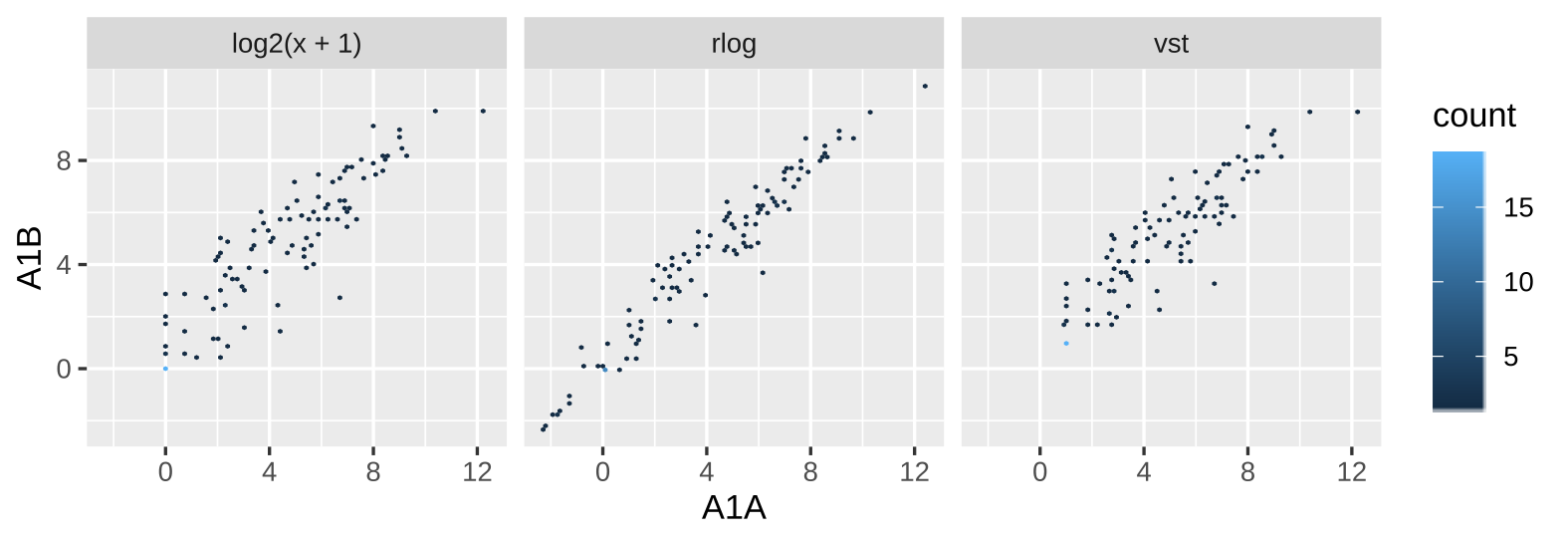 ] .panel[ .panel-name[ R Code ] ```r targetspath <- system.file("extdata", "targets.txt", package = "systemPipeR") targets <- read.delim(targetspath, comment = "#") cmp <- systemPipeR::readComp(file = targetspath, format = "matrix", delim = "-") countMatrixPath <- system.file("extdata", "countDFeByg.xls", package = "systemPipeR") countMatrix <- read.delim(countMatrixPath, row.names = 1) exploreDDSplot(countMatrix, targets, cmp = cmp[[1]], preFilter = NULL, samples = c(3, 4), savePlot = TRUE, filePlot = "transf.png") ``` ] ] --- ### <i class="far fa-chart-bar"></i> Heatmap .panelset[ .panel[.panel-name[Samples] <img src="heat_samples.svg" height="425px" class="center" /> ] .panel[.panel-name[Individuals] <img src="heat_ind.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] #### Samples ```r exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="rlog") heatMaplot(exploredds, clust="samples") heatMaplot(exploredds, clust="samples", plotly = TRUE) ``` #### Individuals genes identified in DEG analysis ```r ### DEG analysis with `systemPipeR` degseqDF <- systemPipeR::run_DESeq2(countDF = countMatrix, targets = targets, cmp = cmp[[1]], independent = FALSE) DEG_list <- systemPipeR::filterDEGs(degDF = degseqDF, filter = c(Fold = 2, FDR = 10)) heatMaplot(exploredds, clust="ind", DEGlist = unique(as.character(unlist(DEG_list[[1]])))) heatMaplot(exploredds, clust="ind", DEGlist = unique(as.character(unlist(DEG_list[[1]]))), plotly = TRUE) ``` ]] --- ### <i class="far fa-chart-bar"></i> Dendrogram .panelset[ .panel[.panel-name[Plot] <img src="cor.svg" height="425px" class="center" /> ] .panel[ .panel-name[ R Code ] ```r exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="rlog") hclustplot(exploredds, method = "spearman") hclustplot(exploredds, method = "spearman", savePlot = TRUE, filePlot = "cor.pdf") ``` ] ] --- ### <i class="far fa-chart-bar"></i> PCA plot .panelset[ .panel[.panel-name[Plot plotly] <div id="htmlwidget-56a5ba518973d0773b79" style="width:504px;height:504px;" class="plotly html-widget"></div> <script type="application/json" data-for="htmlwidget-56a5ba518973d0773b79">{"x":{"data":[{"x":[-3.03168381476885,-6.03412691707515],"y":[-0.535073099413187,-2.0971750907225],"text":["colour: A1<br />PC1: -3.0316838<br />PC2: -0.5350731","colour: A1<br />PC1: -6.0341269<br />PC2: -2.0971751"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(248,118,109,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(248,118,109,1)"}},"hoveron":"points","name":"A1","legendgroup":"A1","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[3.88869312311426,-0.383756141678079],"y":[0.0701335027868824,4.53821439195817],"text":["colour: A12<br />PC1: 3.8886931<br />PC2: 0.0701335","colour: A12<br />PC1: -0.3837561<br />PC2: 4.5382144"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(211,146,0,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(211,146,0,1)"}},"hoveron":"points","name":"A12","legendgroup":"A12","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[3.28184331604165,-0.416809087724184],"y":[0.185372641000547,5.30010399483779],"text":["colour: A6<br />PC1: 3.2818433<br />PC2: 0.1853726","colour: A6<br />PC1: -0.4168091<br />PC2: 5.3001040"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(147,170,0,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(147,170,0,1)"}},"hoveron":"points","name":"A6","legendgroup":"A6","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[-4.23478929449673,-5.72288504075975],"y":[2.46441944072604,-1.3362445127839],"text":["colour: M1<br />PC1: -4.2347893<br />PC2: 2.4644194","colour: M1<br />PC1: -5.7228850<br />PC2: -1.3362445"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,186,56,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(0,186,56,1)"}},"hoveron":"points","name":"M1","legendgroup":"M1","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[3.08219911545694,4.0916538760623],"y":[-1.26931397709069,-3.52388688937858],"text":["colour: M12<br />PC1: 3.0821991<br />PC2: -1.2693140","colour: M12<br />PC1: 4.0916539<br />PC2: -3.5238869"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,193,159,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(0,193,159,1)"}},"hoveron":"points","name":"M12","legendgroup":"M12","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2.40175386965523,1.53523865252027],"y":[1.67449958551854,0.392038639964049],"text":["colour: M6<br />PC1: 2.4017539<br />PC2: 1.6744996","colour: M6<br />PC1: 1.5352387<br />PC2: 0.3920386"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(0,185,227,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(0,185,227,1)"}},"hoveron":"points","name":"M6","legendgroup":"M6","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[-4.10114199086997,-5.13397514161901],"y":[-0.524772724724376,-2.59419479868824],"text":["colour: V1<br />PC1: -4.1011420<br />PC2: -0.5247727","colour: V1<br />PC1: -5.1339751<br />PC2: -2.5941948"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(97,156,255,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(97,156,255,1)"}},"hoveron":"points","name":"V1","legendgroup":"V1","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[2.70779870992001,1.85669227563915],"y":[-0.325101355885941,1.77346995606566],"text":["colour: V12<br />PC1: 2.7077987<br />PC2: -0.3251014","colour: V12<br />PC1: 1.8566923<br />PC2: 1.7734700"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(219,114,251,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(219,114,251,1)"}},"hoveron":"points","name":"V12","legendgroup":"V12","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null},{"x":[3.29098889669914,2.92230559388278],"y":[-0.928463576912153,-3.26402612725811],"text":["colour: V6<br />PC1: 3.2909889<br />PC2: -0.9284636","colour: V6<br />PC1: 2.9223056<br />PC2: -3.2640261"],"type":"scatter","mode":"markers","marker":{"autocolorscale":false,"color":"rgba(255,97,195,1)","opacity":1,"size":11.3385826771654,"symbol":"circle","line":{"width":1.88976377952756,"color":"rgba(255,97,195,1)"}},"hoveron":"points","name":"V6","legendgroup":"V6","showlegend":true,"xaxis":"x","yaxis":"y","hoverinfo":"text","frame":null}],"layout":{"margin":{"t":40.8401826484018,"r":7.30593607305936,"b":37.2602739726027,"l":37.2602739726027},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187},"title":{"text":"Principal Component Analysis (PCA)","font":{"color":"rgba(0,0,0,1)","family":"","size":17.5342465753425},"x":0,"xref":"paper"},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-6.54041595673202,4.59794291571917],"tickmode":"array","ticktext":["-6","-4","-2","0","2","4"],"tickvals":[-6,-4,-2,0,2,4],"categoryorder":"array","categoryarray":["-6","-4","-2","0","2","4"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"y","title":{"text":"PC1: 39% variance","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"scaleanchor":"y","scaleratio":1,"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-3.9650864335894,5.74130353904861],"tickmode":"array","ticktext":["-2","0","2","4"],"tickvals":[-2,0,2,4],"categoryorder":"array","categoryarray":["-2","0","2","4"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"x","title":{"text":"PC2: 17% variance","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"scaleanchor":"x","scaleratio":1,"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":true,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.88976377952756,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.689497716895},"y":0.938132733408324},"annotations":[{"text":"colour","x":1.02,"y":1,"showarrow":false,"ax":0,"ay":0,"font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187},"xref":"paper","yref":"paper","textangle":-0,"xanchor":"left","yanchor":"bottom","legendTitle":true}],"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","showSendToCloud":false},"source":"A","attrs":{"3a05f57e01ae":{"colour":{},"x":{},"y":{},"type":"scatter"}},"cur_data":"3a05f57e01ae","visdat":{"3a05f57e01ae":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> ] .panel[.panel-name[Plot ggplot2] <img src="pca.svg" height="425px" class="center" /> ] .panel[ .panel-name[ R Code ] ```r library(systemPipeShiny) targetspath <- system.file("extdata", "targets.txt", package="systemPipeR") targets <- read.delim(targetspath, comment="#") cmp <- systemPipeR::readComp(file=targetspath, format="matrix", delim="-") countMatrixPath <- system.file("extdata", "countDFeByg.xls", package="systemPipeR") countMatrix <- read.delim(countMatrixPath, row.names=1) exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="rlog") PCAplot(exploredds, plotly = TRUE) ``` ] ] --- ### <i class="far fa-chart-bar"></i> Multidimensional Scaling (MDS) plot .panelset[ .panel[.panel-name[Plot] <img src="mds.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="rlog") MDSplot(exploredds, plotly = FALSE) ``` ]] --- ### <i class="far fa-chart-bar"></i> Generalized Principal Components Analysis .panelset[ .panel[.panel-name[Plot] <img src="glm.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="raw") GLMplot(exploredds, plotly = FALSE) GLMplot(exploredds, plotly = FALSE, savePlot = TRUE, filePlot = "GML.pdf") ``` ] ] --- ### <i class="far fa-chart-bar"></i> Bland–Altman Plot (MA-Plot) .panelset[ .panel[.panel-name[Plot] <img src="maplot.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r exploredds <- exploreDDS(countMatrix, targets, cmp=cmp[[1]], preFilter=NULL, transformationMethod="raw") GLMplot(exploredds, plotly = FALSE) GLMplot(exploredds, plotly = FALSE, savePlot = TRUE, filePlot = "GML.pdf") ``` ] ] --- ### <i class="far fa-chart-bar"></i> t-SNE Plot .panelset[ .panel[.panel-name[Plot] <img src="tsne.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r targetspath <- system.file("extdata", "targets.txt", package="systemPipeR") targets <- read.delim(targetspath, comment="#") cmp <- systemPipeR::readComp(file=targetspath, format="matrix", delim="-") countMatrixPath <- system.file("extdata", "countDFeByg.xls", package="systemPipeR") countMatrix <- read.delim(countMatrixPath, row.names=1) set.seed(42) ## Set a seed if you want reproducible results tSNEplot(countMatrix, targets, perplexity = 5) ``` ] ] --- ### <i class="far fa-chart-bar"></i> Volcano Plot .panelset[ .panel[.panel-name[Plot] <img src="volcano.svg" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r degseqDF <- systemPipeR::run_DESeq2(countDF = countMatrix, targets = targets, cmp = cmp[[1]], independent = FALSE) DEG_list <- systemPipeR::filterDEGs(degDF = degseqDF, filter = c(Fold = 2, FDR = 10)) volcanoplot(degseqDF, comparison = "M12-A12", filter = c(Fold = 2, FDR = 10)) volcanoplot(degseqDF, comparison = "M12-A12", filter = c(Fold = 1, FDR = 20), genes = "ATCG00280") ``` ] ] --- class: inverse, center, middle # <i class="fas fa-code"></i> Customize and Extend --- ## <i <i class="far fa-chart-bar"></i> Development of New Tabs Create new template tabs with `newTabData` and `newTabPlot`. ```r newTabData( tab_id = "data_new", tab_displayname = "my first data tab", prepro_methods = list(makePrepro(label = "do nothing", plot_options = "plot_new")) ) newTabPlot( tab_id = "plot_new", tab_displayname = "my first plot tab", plot_data = list(makePlotData(dataset_label = "Data from my new tab", receive_datatab_ids = "data_new")) ) ``` --- ## <i class="far fa-folder-open"></i> SPS Folder Structure Directory structure with the new tabs: ``` SPS_YYYYMMDD ├── server.R ├── ui.R ├── global.R ## Requires actions! ├── deploy.R ├── config/ │ ├── sps.db │ ├── sps_options.yaml │ └── tabs.csv ## It will be automatically edit ├── R/ │ ├── tab_vs_data_new.R │ └── tabs_vs_plot_new.R ├── data/ │ └── inputData ├── results/ │ └── plot_xx.png └── www/ ``` --- ## <i class="far fa-chart-bar"></i> Development of New Tabs Add new tabs on `global.R` file: ```r sps_app <- sps( vstabs = c("data_new", "plot_new"), # add new tab IDs here server_expr = { msg("Custom expression runs -- Hello World: checking", "GREETING", "green") } ) ``` Launch the app again: ```r shiny::runApp() ``` --- ## <i class="fas fa-users"></i> systemPipeR Team <br/> <table> <tr> <td width="25%"><a href="https://github.com/dcassol"><img src="https://avatars1.githubusercontent.com/u/12722576?s=400&u=092906a3361e7c736e10e3ffab7b60c436cbc56d&v=4"></a></td> <td width="25%"><a href="https://github.com/lz100"><img src="https://avatars2.githubusercontent.com/u/35240440?s=400&u=7dc9f767799cb4f6a1cb584130acc9938f089146&v=4"></a></td> <td width="25%"><a href="https://github.com/mathrj"><img src="https://avatars2.githubusercontent.com/u/45085174?s=400&u=3b9794f20c54948504a261fc7c9c17bdda3a5e1f&v=4"></a></td> <td width="25%"><a href="https://github.com/tgirke"><img src="https://avatars0.githubusercontent.com/u/1336916?s=400&u=8fa9ede9f2f28607ce9e0db83ac1b5b50d2357c7&v=4"></a></td> </tr> <tr> <td align='center' width="25%">Daniela Cassol</td> <td align='center' width="25%">Le Zhang</td> <td align='center' width="25%">Ponmathi Ramasamy</td> <td align='center' width="25%">Thomas Girke</td> </tr> </table> --- class: middle # Thanks! <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Browse source code at <a href="https://github.com/systemPipeR/systemPipeShiny/"> <i class="fab fa-github fa-2x"></i></a> <i class="fas fa-hand-point-right" style="color:#00758a;"></i> Ask a question about systemPipeShiny at Bioconductor Support Page <a href="https://support.bioconductor.org/"><i class="far fa-question-circle fa-2x"></i></a> <i class="fas fa-hand-point-right" style="color:#00758a;"></i> **systemPipeR** at [Bioconductor](http://www.bioconductor.org/packages/release/bioc/html/systemPipeR.html) <i class="fas fa-hand-point-right" style="color:#00758a;"></i> [https://systempipe.org/](https://systempipe.org/) --- class: inverse, center, middle # <i class="fas fa-list"></i> ToDo List --- ## <i class="fas fa-list"></i> Doing - Documentation: *Vignette* - Complete the visualizations tabs with the plots controls - Unit Testing - Add Quality plots - Add Functional Enrichment Analysis plots - Final deployment (shinyapp.io) and Review package - Submission deadline --- ### <i class="far fa-chart-bar"></i> Functional Enrichment Analysis plots .panelset[ .panel[.panel-name[Dotplot] <img src="dotplot.png" height="425px" class="center" /> ] .panel[.panel-name[Encrichment Map] <img src="emapplot.png" height="425px" class="center" /> ] .panel[.panel-name[Category Netplot] <img src="cnetplot.png" height="425px" class="center" /> ] .panel[.panel-name[Ridgeplot] 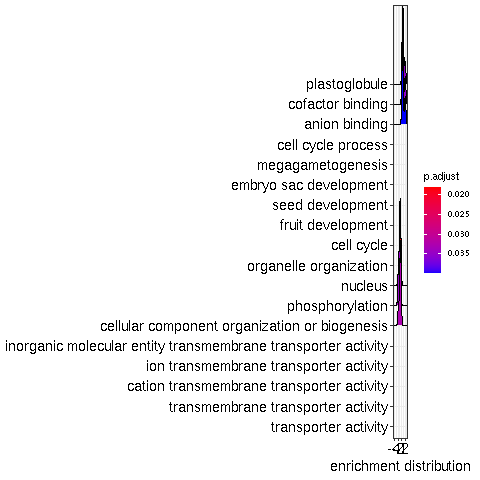 ] .panel[.panel-name[GSEA Plot] <img src="gseaplot.png" height="425px" class="center" /> ] .panel[ .panel-name[R Code] ```r ## Still working ``` ] ]